8.11 EMP_network_plot
The EMP_network_plot module can visualize the results of network analysis in graphical form. This section can be referenced together with Section 6.12.
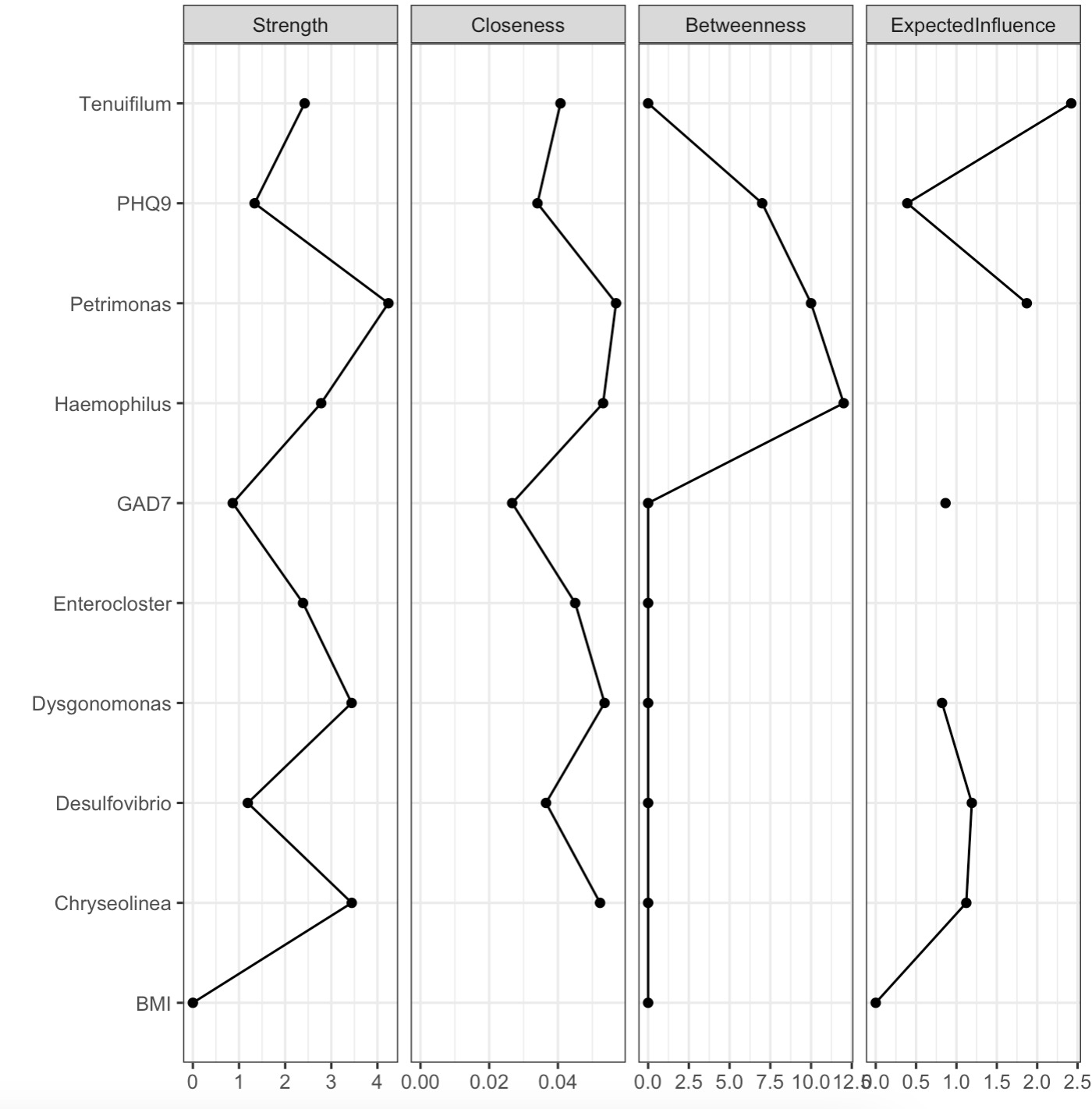
8.11.1 Visualization of note importance
Note:
Parameter
Parameter
show could show the net or node。🏷️Example:
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_collapse(estimate_group = 'Genus',collapse_by = 'row') |>
EMP_diff_analysis(method='wilcox.test', estimate_group = 'Group') |>
EMP_filter(feature_condition = pvalue<0.05) |>
EMP_network_analysis(coldata_to_assay = c('BMI','PHQ9','GAD7')) |>
EMP_network_plot(show='node')
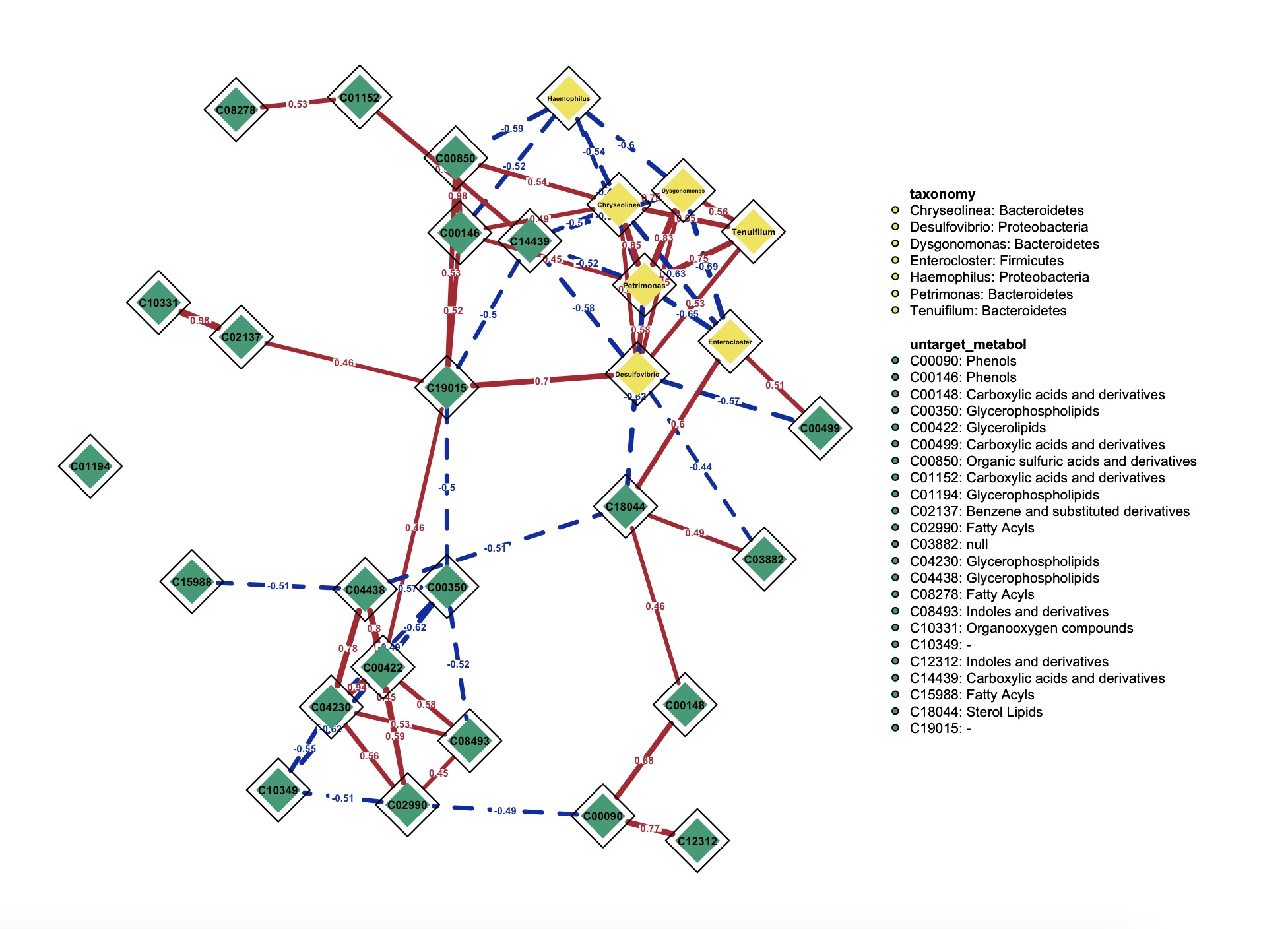
8.11.2 Visualization of network
Note:
This module inherits the parameter usage from qgraph.
This module inherits the parameter usage from qgraph.
🏷️Example:
k1 <- MAE |>
EMP_assay_extract('taxonomy') |>
EMP_collapse(estimate_group = 'Genus',collapse_by = 'row') |>
EMP_diff_analysis(method='wilcox.test', estimate_group = 'Group') |>
EMP_filter(feature_condition = pvalue<0.05)
k2 <- MAE |>
EMP_collapse(experiment = 'untarget_metabol',na_string=c('NA','null','','-'),
estimate_group = 'MS2kegg',method = 'sum',collapse_by = 'row') |>
EMP_diff_analysis(method='DESeq2', .formula = ~Group) |>
EMP_filter(feature_condition = pvalue<0.05)
(k1 + k2 ) |>
EMP_network_analysis() |>
EMP_network_plot(show = 'net',layout = 'spring',
shape='diamond',
edge.labels=TRUE,edge.label.cex=0.4,
vsize = 5,threshold = 0,
node_info = c('Phylum','MS2class'),
legend.cex=0.3,label.cex = 1,label.prop = 0.9,font=2)